Modeling Molecules
Thanks to the space program, scientists are modeling molecules with the goal of effectively discovering new treatments for diseases. NASA's Goddard Space Flight Center, along with other government organizations, provided Small Business Innovation Research (SBIR) funding for technology that could simulate spacecraft, vehicle, and machine dynamics, as well as biomechanical motions. This SBIR project spunoff into the development of software that can model the dynamics of molecules. Moldyn, Inc., of Cambridge, Massachusetts, patented a technology that can simulate low-frequency molecular motions and properties, such as movements among a molecule's constituent parts.
This commercially available modeling method is known as Multibody Order (N) Dynamics (MBO(N)D). In MBO(N)D, a molecule is substructured into a set of interconnected rigid and flexible bodies. Substructuring organizes groups of similarly behaving atoms into interacting flexible bodies. These flexible bodies replace the computational burden of mapping each individual atom. MBO(N)D reduces the number of calculations from tens of thousands of atoms to a few hundred, or less, bodies. Conventional atom-by-atom molecular dynamics can require weeks, sometimes months, to simulate the behavior of a few molecules. Moldyn's multibody approach cuts the computational time by up to a hundredfold, while also yielding comparable accuracy with all-atom methods.
Moldyn's MBO(N)D technology is available as Insight II 97.0 from Molecular Simulations, Inc. The software is an accelerator for molecular dynamics that uses the same mathematical algorithms originally developed for large flexible satellites to reach time and size scales that were not attainable through conventional methods. In the aerospace industry, the method is used to account for forces on hundreds of thousands of parts in spacecraft and satellites.
Pharmaceutical companies are using the method for a variety of molecular analyses, including realistic modeling of drug/protein interactions and comparison of these interactions with others found in vast databases. Drug companies use databases containing hundreds of thousands to millions of different molecules and molecule fragments to look for potential medicines. The MBO(N)D software allows efficient screening of these large databases to find molecules that can "dock" successfully with the active sites of proteins. The result of this innovation is faster, less expensive drug development than what is allowed by conventional atom-by-atom computational methods.
Drug discovery has changed over the past two decades to include the study of significant structural components in and around the cell. Molecular structures of proteins and receptor-ligand complexes play an important role in the discovery of pharmaceutical agents. A ligand is a biological compound that fits a receptor within or on the cell surface. This fitting is analogous to a lock-and-key arrangement. Drugs that mimic the shape of a natural ligand can be used to regulate a signal in order to manipulate a therapeutic interaction between the synthetic ligand and the cell.
A major breakthrough with this development is to permit the solution of realistic-sized molecular dynamics problems on a moderate workstation, rather than on supercomputers.
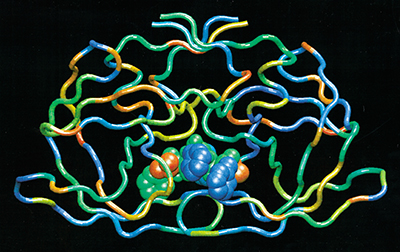
Pharmaceutical companies are using the method for a variety of molecular analyses, including realistic modeling of drug/protein interactions.